November 17, 2006
Premature Journal Club
"A linguistic model for the rational design of antimicrobial peptides" belongs loosely within a respectable research tradition of attempting to understand complex molecular biological systems -- in particular, genes and their products -- in terms of language. Genes encode information and are richly expressive, encompassing (to leading order) the breadth and variation of life on Earth. Moreover, they exhibit both modularity and consistency, to the extent that structures having a particular significance in one location may exhibit the same significance elsewhere. Obviously, the ultimate "interpretation" occurs biophysically, in terms of thermodynamics etc, rather than just by ascription of meaning to arbitrary signifiers, but there is nevertheless at least an intuitive sense in which the encoding must obey some structural rules. As humans, we already have a powerful understanding of rule-based systems for encoding meaning, because we use them all the time -- I'm doing so right now as I write, and so are you as you read. Those systems are languages, and their formal study has given rise to various tools which might conceivably be applied in a biological context. Grammars are systems of rules for managing language symbolically, formally specifying how the symbols -- eg, words -- can legitimately be strung together and with what structural effects. Grammars operate primarily at the level of syntax rather than semantics, as this famous example by Noam Chomsky, the father of modern linguistics, demonstrates:
Colourless green ideas sleep furiously
The sentence is syntactically correct (modulo some reasonable assumptions as to the grammar we're choosing to parse it with), but meaningless. This is a distinction that grammars can't, on the whole, make. (Things, as always, are not quite as clear-cut as this, but let's ignore that for the moment.)
Chomsky defined a famous hierarchy of grammars that is still widely used today, according to the difficulty of either generating or parsing the language. The simplest grammars are Type 3, defining regular languages, allowing only very basic and mechanical local symbolic relationships, while the most complex are Type 0, defining recursively enumerable languages, in which very complex relationships may exist between widely separated parts of a sentence. (Type 0 languages are Turing equivalent in the sense that parsing them requires the same features as universal computation.)
Natural human languages are almost always Type 0, and it is likely that this is also the general case for biological systems. Nevertheless, many subsystems may be more tractable, and this paper takes such an approach with respect to one particular kind of biological sequence.
Antimicrobial peptides are small proteins -- typically of the order of tens of amino acids in length and without extensive secondary or tertiary structures -- that occur widely throughout the eukaryote kingdom as defences against microbial infection. They exploit differences in cell membrane composition between the host organism and the microbes to selectively disable or destroy invaders while not posing a threat to the organism's own cells. These capabilities are static and comparatively cheap, unlike the more complex responses of an adaptive immune system; if the latter exists, it will be in addition to AmP defences rather than as a replacement for them.
One surprising feature of AmPs is that, even though they are very widespread and have apparently been around in at least some form since virtually the dawn of time, they seem unsusceptible to resistance. Possibly their modes of operation are so fundamental that a bacterium would need to transform itself out of all recognition in order to escape, with no noticeable selective advantage for the intermediate states.
Whatever the reason, this offers some hope that AmPs may be a source of therapies that will remain potent in the face of antibiotic-resistant agents. However, there is a corresponding concern that modelling drugs too closely on naturally-occurring AmPs might promote the development of resistance, with potentially catastrophic consequences for defences on which we all depend. One suggested avenue is to try to develop novel AmPs, with similar antimicrobial effects but without close resemblance to any existing ones.
The authors of this paper hypothesize that AmPs can be modelled linguistically. Because they are relatively simple, common and very old, there might be coherent idioms that hold across a wide range of variations in different organisms. Successful AmPs would operate within this linguistic space, and our newfangled ones should too, but occupying portions of the space that have not been significantly colonised so far.
Their experiment is (conceptually) simple: extrapolate AmP idioms to create novel peptides, then see if they work.
A large number of AmP sequences are known, and these were analysed using the standard motif-finding program Teiresias. (A motif is just a shared pattern that is common to the set of sequences.) The derived motifs were filtered to include only patterns common to AmPs -- but rare in other non-antimicrobial proteins -- rather than those common everywhere. The resulting motifs were treated as regular grammars.
It's important to note a decision made here: that the terminal symbols of the AmP language will be single amino acid residues. This is conceptually similar to defining an English grammar in terms of letters rather than -- as is usual -- parts of speech. For a human language like English, such an attempt would be doomed to failure. However, we don't really have any higher-level classification of the symbolic units of biological sequences that makes any sense in this context, so there isn't really much choice. Moreover, different amino acids do have different physiochemical characteristics and could thus be seen to carry different meanings, so the approach is somewhat justifiable, despite the overall whiff of desperation.
Having identified about 700 10-residue patterns to be considered grammatical, 20-residue sequences were generated that conformed to at least one of those patterns at every constituent 10-residue window, like this:
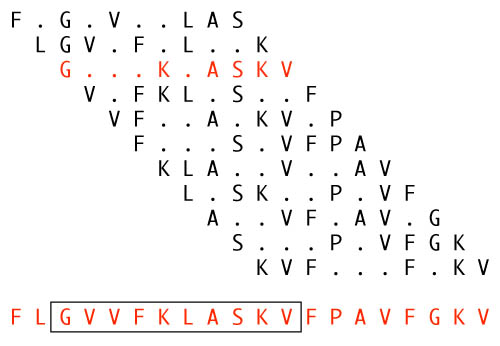 (The 20-mer shown at the bottom is one of the experiment's actual sequences, D28. The contributing patterns were not published, so those shown have just been randomly invented to illustrate the concept.)
To avoid too much similarity with real AmPs, sequences with six or more consecutive amino acids the same as a known AmP sequence were discarded. Those remaining were ranked according to the popularity of the matched grammars, and a bunch of promising sequences were synthesised. These were tested for antimicrobial effects alongside known-good AmPs, known non-AmPs and other artificial sequences containing the exact same amino acids but in ungrammatical order.
The results were reasonably promising: nearly half the "designed" peptides showed antimicrobial effects, and some were competitive with naturally-occurring AmPs. The shuffled versions, by contrast, were almost all ineffective, demonstrating the truism that primary structure is fundamental to protein behaviour.
In this particular case, it appears that even a simplistic language model has some merit, although it clearly doesn't constitute any kind of decoding -- it is more like one of those web toys that concoct movie plots or advertising slogans or whatever by stringing together random words and phrases from predefined pools in some templated manner:
(The 20-mer shown at the bottom is one of the experiment's actual sequences, D28. The contributing patterns were not published, so those shown have just been randomly invented to illustrate the concept.)
To avoid too much similarity with real AmPs, sequences with six or more consecutive amino acids the same as a known AmP sequence were discarded. Those remaining were ranked according to the popularity of the matched grammars, and a bunch of promising sequences were synthesised. These were tested for antimicrobial effects alongside known-good AmPs, known non-AmPs and other artificial sequences containing the exact same amino acids but in ungrammatical order.
The results were reasonably promising: nearly half the "designed" peptides showed antimicrobial effects, and some were competitive with naturally-occurring AmPs. The shuffled versions, by contrast, were almost all ineffective, demonstrating the truism that primary structure is fundamental to protein behaviour.
In this particular case, it appears that even a simplistic language model has some merit, although it clearly doesn't constitute any kind of decoding -- it is more like one of those web toys that concoct movie plots or advertising slogans or whatever by stringing together random words and phrases from predefined pools in some templated manner:
He was a telephone sanitiser from the wrong side of the tracks. She was a bottle-nosed dolphin. Together they would crack the greatest secret known to washing machines!
Such an approach clearly has little or no applicability beyond the particular domain to which it is fitted, and only very constrained domains are likely to be susceptible to such fitting; nevertheless, where it works it may serve as a valuable heuristic for pruning a combinatorially-explosive problem space to the point where workable solutions can be found.
Posted by matt at November 17, 2006 01:30 PM
Comments
Something to say? Click here.